This project supports novel computer science research to provide foundations for new technologies. The research objective of this project is to design new robust data mining and machine learning algorithms for solving the computational challenges in complex materials genome data mining. The Materials Genome Initiative research has been launched by U.S. government to discover, manufacture, and deploy advanced materials fast and low-cost, which holds great opportunities to address the challenges in clean energy, national security, and human welfare. However, the major computational challenges are the bottlenecks for comprehensive materials genome data analysis due to unprecedented scale and complexity. There is a critical need for new data mining and machine learning strategies to bridge the gap and facilitate the new materials discovery. To solve the key and challenging problems in mining such comprehensive heterogeneous materials genome data, this project is to develop a novel robust data mining and explore ways to integrate features from multiple data sources.
Scientific outcomes for intellectual merits:
- We developed multiple sparse multi-view learning algorithms for identifying synthesis markers related to quantitative and qualitative catalytic performance of synthesized nanoparticles over time.
- Data integration to combine the numerical descriptions from different aspects of the nanoparticles generated by different material characterization methods become imperative to utilize the full potential of all available data. New sparse multi-task multi-view feature selection methods were developed to integrate heterogeneous data and identify the informative features to improve both catalytic capability prediction and association between material characterization and theoretical modeling measures. These identified features are then studied together over time to uncover temporal relationship between the longitudinal characterization measures and progressive catalytic performances of synthesized nanoparticles (See Figure 1).
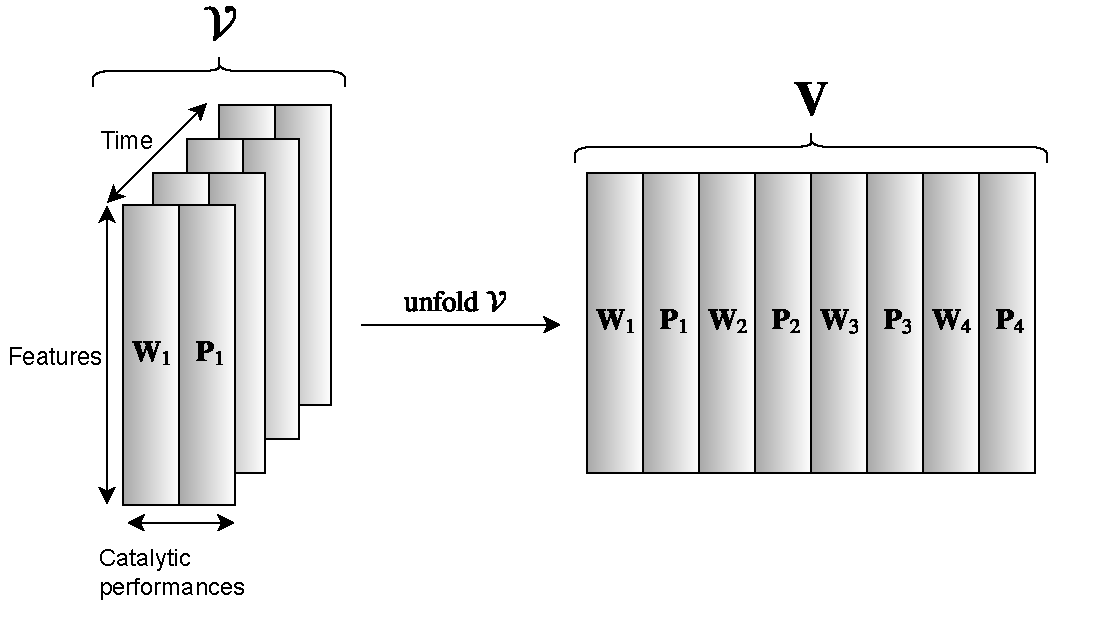
Figure 1: This is designed to find catalytic correlations that are consistent over time and correspond to the modal structure of input characterization data.
-
We developed semi-supervised classifications algorithm via elastic and robust embedding (ERE) for catalytic capability prediction of synthesized nanoparticles. Transductive semi-supervised learning can only predict labels for unlabeled data appearing in training data, and cannot predict labels for testing data never appearing in training set. To handle this out-of-sample problem, many inductive methods make a constraint such that the predicted label matrix should be exactly equal to a linear model. In practice, this constraint might be too rigid to capture the manifold structure of data. We relaxed this rigid constraint and developed a model to use an elastic constraint on the predicted label matrix such that the manifold structure can be better explored. Moreover, since unlabeled data are often very abundant in practice and usually there are some outliers, we used a non-squared loss instead of the traditional squared loss to learn a robust model.
-
We developed a multi-instance enriched data representation algorithm for characterizing nanoparticle data at different granularity levels (See figure 2). Multi-instance learning (MIL) has demonstrated its usefulness in many real-world image applications in recent years. However, two critical challenges prevent one from effectively using MIL in practice. First, existing MIL methods routinely model the predictive targets using the instances of input bags, but rarely utilize an input bag as a whole. As a result, the useful information conveyed by the holistic representation of an input bag could be potentially lost. Second, the varied numbers of the instances of the input bags in a data set make it infeasible to use traditional learning models that can only deal with single-vector inputs. To tackle these two challenges, we developed a novel multi-instance representation learning method that can integrate the instances of an input bag and its holistic representation into one single-vector representation.
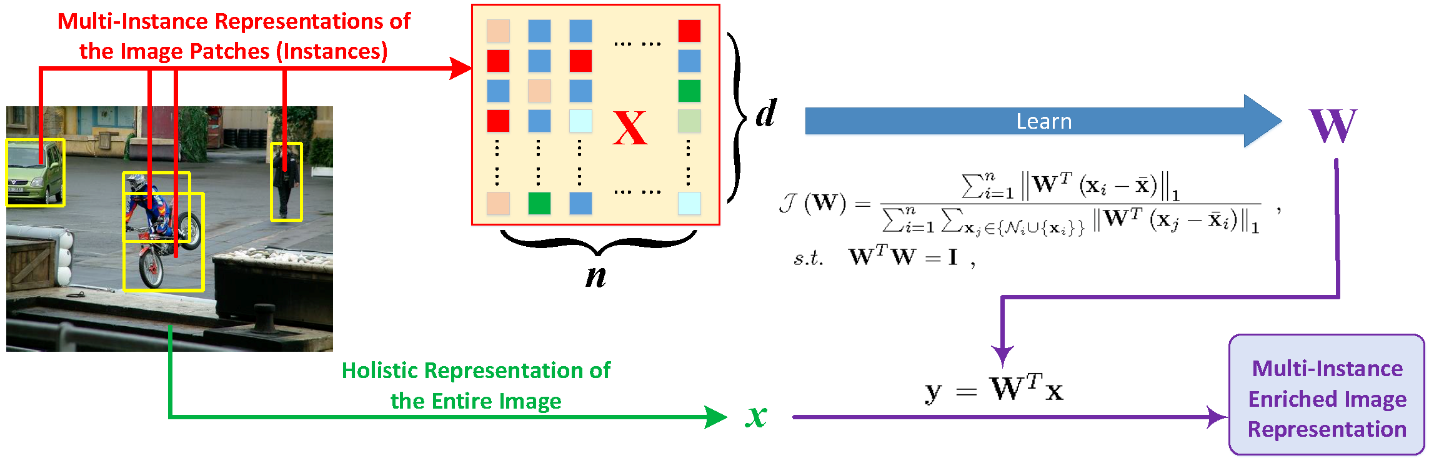
Figure 2: We use images to illustrate the representation learning for multi-instance data where an image is a bag and the image patches are instances. This picture is in our IEEE CVPR 2018 paper.
-
We have performed analysis on Identifying the enzymatic mode of action for cellulase enzymes via docking calculations using developed machine learning algorithms. The binding distributions for cellobiose have been classified into two distinct types; distributions with a single maximum or distributions with a bi-modal structure. It is found that the uni-modal distributions correspond to exo- type enzyme while a bi-modal substrate docking distribution corresponds to endo- type enzyme. These results indicate that the use of docking calculations and machine learning algorithms are a fast and computationally inexpensive method for predicting if a cellulase enzyme possesses primarily endo- or exo- type behavior, while also revealing critical enzyme-substrate interactions (see Figure. 3).
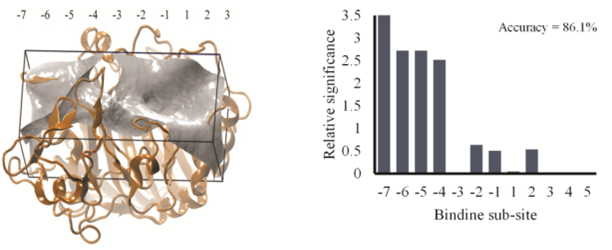
Figure 3: Machine learning algorithm results for the carboxymethyl cellulose docking calculation. A larger relative significance indicates a larger favorable contribution to the predictive model.
Other outcomes for broader impacts:
- We have published about 20 full-length papers related to this project in peer-reviewed conference proceedings and journals.
- This project supported two Ph.D. students at Colorado School of Mines. One of them have graduated and the other one is currently a fifth year Ph.D. student in the Department of Computer Science and will graduate in next year with looking for an academic position.
- This project also supported two undergraduate REU students. The work from one of these two students (together with his graduate student mentor supported by this project) has led to a manuscript submitted to a top-tier peer-reviewed journal.
- The research materials produced in this project are used in teaching several undergraduate and graduate courses at Colorado School of Mines.
