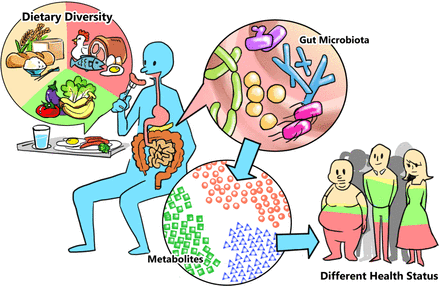
Although communities of microorganisms, originally referred to as microbiota, have been studied for a long time, the field has taken off in 2002 with the advent of metagenomics, which for the first time equipped us with a way to “see” the incredible diversity of species around us — too small to see with or own eyes. Now, the microbiome is not only of interest in environmental samples, but also within our own bodies. Particularly, the gut microbiome has skyrocketed as a source for potential disease biomarkers and treatment options, not only with drastically altered microbiomes in obese individuals, but also in patients with cancer, irritable bowel syndrome, diabetes, asthma, and many others.
Current microbiome data analysis is restricted to identifying differences in representation of organisms in healthy and disease states, primarily using sequence alignments. These approaches are not sufficient for researchers, industries, policy makers, and members of the public to locate, assemble, and integrate data from multiple heterogeneous data sources that meet their needs, especially in informing product formulation and use. This project is to use sophisticated and state-of-the-art machine learning approaches to integrate the microbiome data, extract the information that more directly relates the sequence data to specific outcomes, and ultimately answer specific commercially relevant questions that can move our intellectual properties closer to marketability.